General research interest:
Our research activity is focused on the general aspects of protein structure, structural dynamics and conformational flexibility. We are particularly interested in the role of protein-protein interactions and allosteric conformational transitions in different biological processes. Our model proteins are mainly allosteric enzymes that can self organize to form sophisticated multi-molecular structures. In our research we apply both in silico modeling and wet laboratory experiments in a concerted manner.
Major research projects:
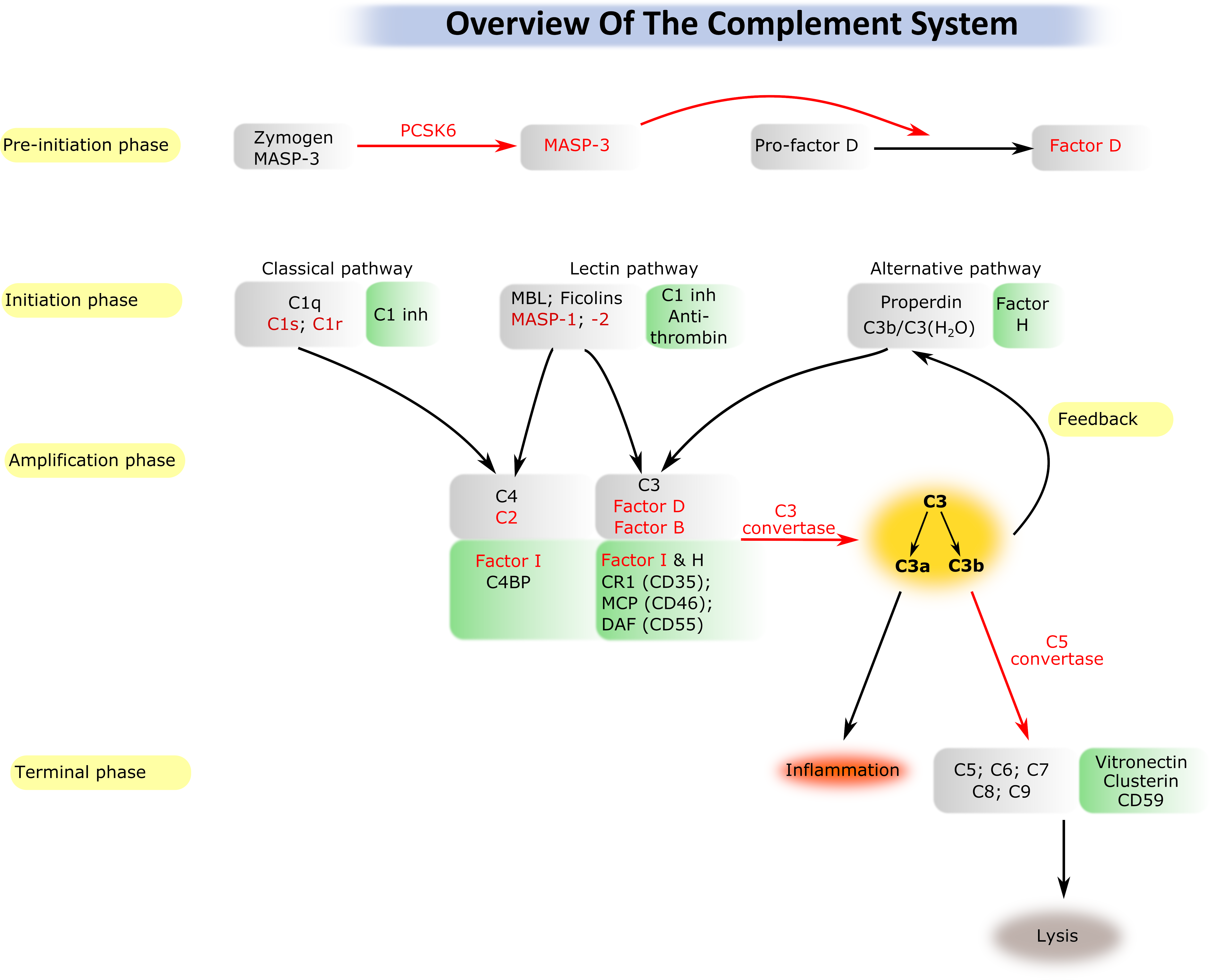
1.) Activation and regulation of the complement system
The complement system (CS) is a major effector arm of the innate immune system. The CS consists of about 40 protein molecules and it is capable of recognizing, labeling and eliminating dangerous structures such as pathogenmicroorganisms. The majonents of the CS are serine protease enzymes that activate each-other in a cascade-like manner. An intact complement system is essential for maintaining the immune homeostasis of the body; however, its uncontrolled, pathological activation may results in self-tissue damage.Recently it has been turned out that the pathological complement activation contributes to the development of numerous disease conditions. Our research concentrates on the study of the early serine proteases that initiate the complement activation. We recombinantly express the proteases in different expression systems and characterize their structural, biochemical and enzymological properties. Special emphasis is laid on the study of the natural and artificial inhibitors of the complement serine proteases. During the recent years we have determined the 3D structure of several serine proteases, we have clarified the exact mechanism of the activation of the lectin pathway of CS, and we have discovered a novel mechanism for initiation of inflammation.
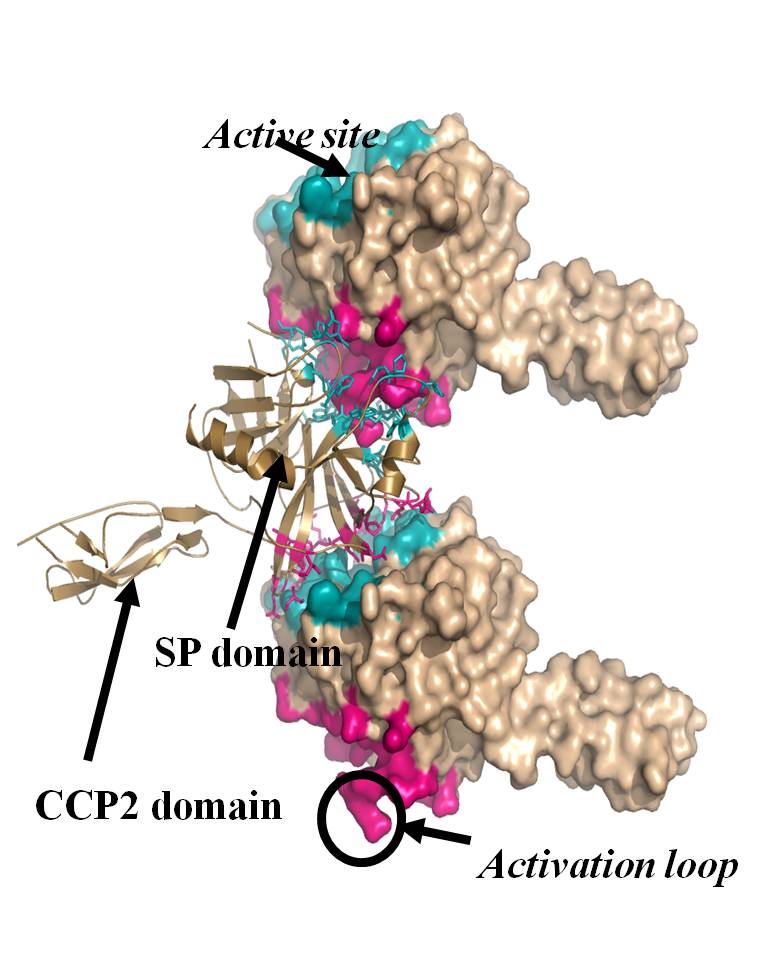
Fig. 1. Crystal structure of autoactivating MASP-2 moleculesment activation contributes to the development of numerous disease conditions.
2.) The functional landscape of proteins
The functional landscape of a protein is the part of its free energy landscape that corresponds to the native state. The functional motions of proteins (catalysis, ligand binding, allosteric regulation) occur on this hypersurface having a complex topology. We plan to reveal the general properties of this landscape (hierarchical structure, the nature of
dynamics, etc.) on a structurally representative subset of proteins, focusing on proteins that are studied in the institute by other methods. We determine the functional dynamics of each protein by performing simulations on models of various resolutions and applying a range of analysis methods. The functional landscape is particularly important for understanding the mechanisms of allostery and allosteric communication pathways. We construct structural models of protein-protein complexes, and describe allosteric communication within these complexes using network models.

Fig.2. Illustration of the free energy landscape of a protein. The functional landscape is the bottom part of the funnel-shaped surface, and it has a complex topology.
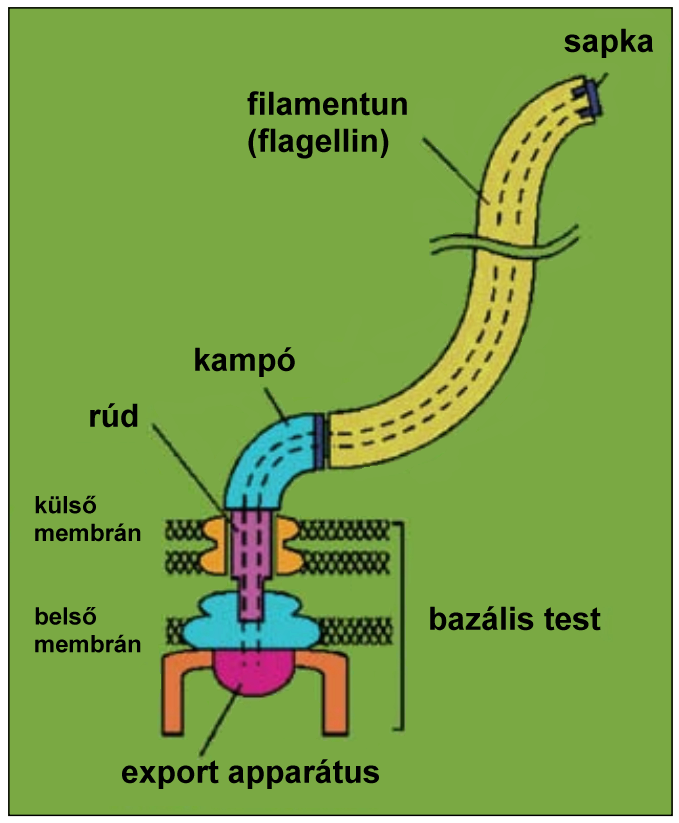
3.) The bacterial flagellar export system
The bacterial flagellum is the main locomotion organelle of bacteria. It is composed of three major parts: the basal body, the hook, and the filament. The filament is basically made of more than ten thousand copies of a single protein,
flagellin. Flagellin (FliC) monomers are transported through a narrow channel of about 20 Å to the tip of the filament where they self-assemble into the growing structure. Flagellin and several other components of the flagellum are transferred to the central channel by the flagellar type III secretion system, or in other words export system. The precise mechanism of this process is unknown. Our focus is to reveal the exact functions of the components of the export system, and furthermore to utilize the flagellar export system to produce recombinant proteins secreted into the medium.
4.) Identification and characterization of interactions between proteins participating in signal transduction processes leading to cancer or inflammation
The essential physiological processes of the living cells are based on the sophisticated network of biological macromolecules, mainly that of proteins. The signal transduction pathways are mediated by different protein-protein interactions (e.g. strong or weak; direct or indirect), that regulate the functions of the allosteric enzymes.
The allosteric regulation, based on the conformational flexibility of proteins, is of great significance in the function of enzymes; and we have previously gained insight into such regulatory mechanisms by using isolated proteins. Based on our knowledge, in the frame of the present project, we would like to identify and characterize enzyme-enzyme interactions at the molecular level that are involved in signal transduction processes having pathophysiological relevance. According to our hypothesis there is a relationship between chronic inflammation and cancer, and in this context we would like to study the role of protein-protein interactions in which certain kinases (e.g. Aurora, Rock1, Rock2 , phosphoglycerate kinase) and phosphodiesterases (e. g. PDE4, PDE5) might be involved. The molecular interactions will be studied in vivo using mammalian cell cultures and in vitro applying enzymological and biochemical methods.
Collaborations
Complement research:
Department of Biochemistry, Department of Chemistry, Eötvös University, Budapest
3rd Department of Internal Medicine, Semmelweis University , Budapest
University of Aarhus, Denmark
University Hospital, Bern, Switzerland
University of British Columbia, Vancouver, Canada
Bioinformatics:
Department of Medical Chemistry, Molecular Biology and Pathobiochemistry , Semmelweis University, Budapest
University of Michigan, USA
Bacterial flagellins:
Pannon University, Veszprém
Instuments
FPLC for protein purification
QCM, DSC, FT IR
Teaching
Undergraduate courses: Pázmány Péter Catholic University, Faculty of Information Technology and Bionics: biophysics, biochemistry and molecular biology
Supervising B.Sc., M.Sc. and Ph.D. students at the Eötvös University and University of Technology
Leader
Péter Gál