Physiological consequences and potential role of uracil substitution in genomes of model organisms
Conventionally, deoxyuridine incorporation into DNA is regarded to represent erroneous lesions, however chemical features of the uracil base does not show a remarkable difference from thymine except from a single methyl group at the 5th position of the pyrimidine ring. As the balance of the cellular nucleotide pool is deterministic in the quality of DNA synthesis, dUTPase catalyzing the hydrolysis of dUTP is a major player in the maintenance of this balance and uracil-free genome. Our research is focused on the genome metabolism of uracil substituted DNA in the framework of a new paradigm, suggesting that deoxyuridine lesions might assign unique fate for DNA in special cases. These cases are extensively studied recently by a set of research groups including ours. The special instances in which the presence of uracil was verified to have a unique role are the following:
- Immunoglobuline gene diversification
- Transcriptional regulation
- HIV life cycle
- and developmental biology
We focus on some of the above mentioned or other examples of uracil-DNA mediated cellular events. We employ this paradigm on model organisms such as mammalian or tumor-derived cell lines and Drosophila melanogaster. The physiological role of uracil substituted DNA is studied either genome scale by methods of molecular and cell biology. In Drosophila, complex developmental consequences are also examined in our lab.


Muha et al, 2012 PLoS Genetics
Interconnections of thymine-less cell death with cellular signaling pathways
Almost all organisms employ a cluster of metabolic enzymes devoted for thymine biosynthesis in order to utilize thymine bases instead of uracil in their genome. The medical significance of this metabolic pathway is marked by the fact that nearly one-third of anti-cancer drugs used in clinics are targeted against thymidilate biosynthesis (such as fluoro-pyrimidines, anti folates) potentially inducing the so called thymine-less cell death. Personalized medicine aiming to minimize side effects and maximize the efficiency of chemotherapies requires prior knowledge about the characteristics of tumorous cells in order to predict the desired effect of a particular drug. Detailed mapping of protein networks participating or being affected by thymine-less cell death help us to estimate the receptivity of tumorous cells for drugs targeting thymidilate biosynthesis. We would like to contribute for this knowledge through our research that includes the characterization of thymine-less cell death. We especially aim to explore the involvement of dUTPase in the cellular process of thymine-less cell death.
Study of a molecular switch: Structural and molecular biological research on the Staphylococcus aureus pathogenicity island regulation
The bacterial genom frequently contains mobile genetic elements, which can replicate more or less independently from the bacterial chromosome. Some of these are phage related such as the pathogenicity islands (PIs), which have significant biomedical importance, since these are responsible for horizontal transfer of several toxins and virulence factors (for eg. the toxic shock syndrome).
Phage mediated mobilisation of some Staphylococcus aureus PIs are induced by formation of a repressor:derepressor complex of the Staphylococcal repressor protein (Stl) with a phage-related dUTPase. Studying the detailed mechanism of this interaction can provide much needed deep insight into bacterial gene expression regulation pathways, and potentially facilitates the design of new anti-bacterial compounds. In this project we investigate this system using various in vitro techniques (native gel electrophoresis, electrophoretic mobility shift assay, steady-state and transient kinetics, VIS and fluorescence spectroscopy, mass spectrometry and X-ray crystallography).
Studying regulation of macromolecular transport between nucleus and cytoplasm in space and time

Nucleotide metabolism pathways important in so-called preventive DNA repair, among these nuclear and mitochondrial localization of dUTPase enzyme in eukaryotes, can universally be observed. Mechanism of the localization and regulation, we would like to explore, is questionable. Apropos of this, we found out the intriguing dynamic regulation role of cell cycle coupled (cdk1 kinase mediated) phosphorylation, that also applies to several proteins of human proteome. Cessation of phosphorylation potential and mimication of constitutive phosphorylation by mutation of serine position to glutamine or glutamic acid decisively affect dUTPase pool of daughter cells’ nuclear proteome. This observation likely reveals a general regulatory mechanism. According to our work hypothesis phosphorylation by cdk1 kinase near NLS significantly influance dynamic nuclear transport of several human proteins.
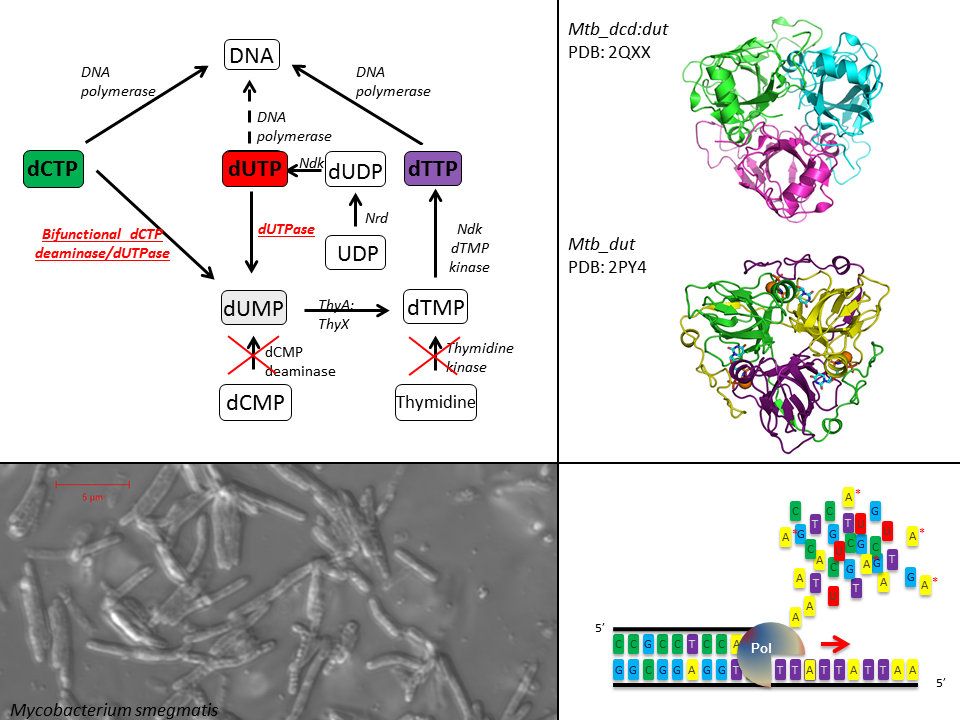
Thymidine biosynthesis is essential as dTTP is one of the nucleotide building blocks of DNA. Normally, three major pathways exist for dTTP synthesis, but only one of these is present in mycobacteria. This one involves the dUTPase reaction which also serves to eliminate excess dUTP to prevent DNA uracilation. Due to the exclusive biosynthetic role of dUTPase in mycobacteria, these organisms are ideal to investigate the physiology of the dUTPase reaction and the mechanisms underlying genom integrity in the living mycobacterial cell. The laboratory model Mycobacterium smegmatis is used for our experiments, where we have successfully rescued the dUTPase knock-out phenotype with several mutant dUTPases. Using a complex array of methods of cell and molecular biology, we are getting novel insights into a biological problem of potentially central interest of cell biology. This system provides unparalleled opportunity to study how mutant dUTPase enzymes perform under normal conditions as well as under diverse shock conditions in the mycobacterial cell.
Structural and mechanistic insights into antimalarial drug target CTP:phosphocholine cytidylyltransferase (CCT) from Plasmodium falciparum
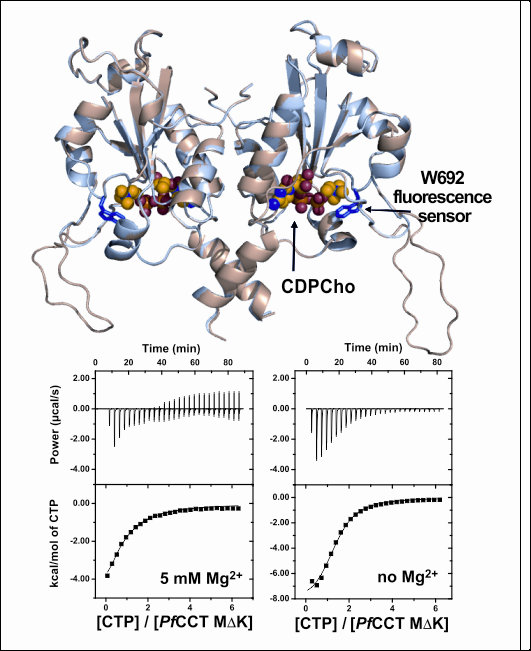
Despite the intensive antimalarial research, malaria is still one of the deadliest infectious diseases today. Amongthe causative agents, Plasmodium falciparum is responsible for the vast majority of mortal outcomes. The recent challenges include the spread of multidrug resistant strains, urging the discovery of novel antimalarial targets. CTP:phosphocholine cytidylyltransferase (CCT) catalyzes the rate-limiting step within de novo phosphatidylcholine biosynthesis, a novel validated antimalarial drug target. We perform biochemical and structural characterization of CCT from Plasmodium falciparum to gain insights in mechanism of action and characterize parasite-specific functional elements.
Current collaborations:
Buday László, MTA TTK
Liliom Károly, MTA TTK
Szüts Dávid, MTA TTK
Vékey Károly, MTA TTK
Perczel András, ELTE
Harmat Veronika, ELTE
Náray-Szabó Gábor, ELTE
Nyitray László, ELTE
Kovács Mihály, ELTE
Málnási-Csizmadia András, ELTE
Vellai Tibor, ELTE
Nyulászi László, BME
Poppe László, BME
Oláh Julianna, BME
Fésüs László, DE
Matthias Wilmanns, EMBL Hamburg
Barabás Orsolya, EMBL Heidelberg
Dolores Gonzalez-Pacanowska, Granada
Keith Wilson, Univ York
Iva Pichova, Czech Academy of Science, Prága
Henri Vial Univ Montpellier
Rachel Cerdan Univ Montpellier
Bostjan Kobe Univ Queensland

Leader
Beáta G. Vértessy


