Signal Transduction and Functional Genomics Research Group
Introduction, main research area
Two separate research laboratories are working in the Signal Transduction and Functional Genomics Research Group cooperating in several fields. The main scope of the Signal Transduction Laboratory (leader: László Buday) is to study the pathways of growth factor signal transduction that utilize tyrosine kinase enzymes to regulate cell division and motility. The Functional Genomics Laboratory (leader: László Patthy) focuses on genome annotation in the field of bioinformatics with similar interest in studying the structure and function of certain proteins of major medical importance. Both laboratories participate in the Protein Science and its Applications National Programme (HunProtEx, 2018-1.2.1-NKP-2018-00005), and the Centre for University-Industry Cooperation (FIEK_16-1-2016-0005) led by ELTE.
Synopsis of the main research projects
- The study of Tks proteins
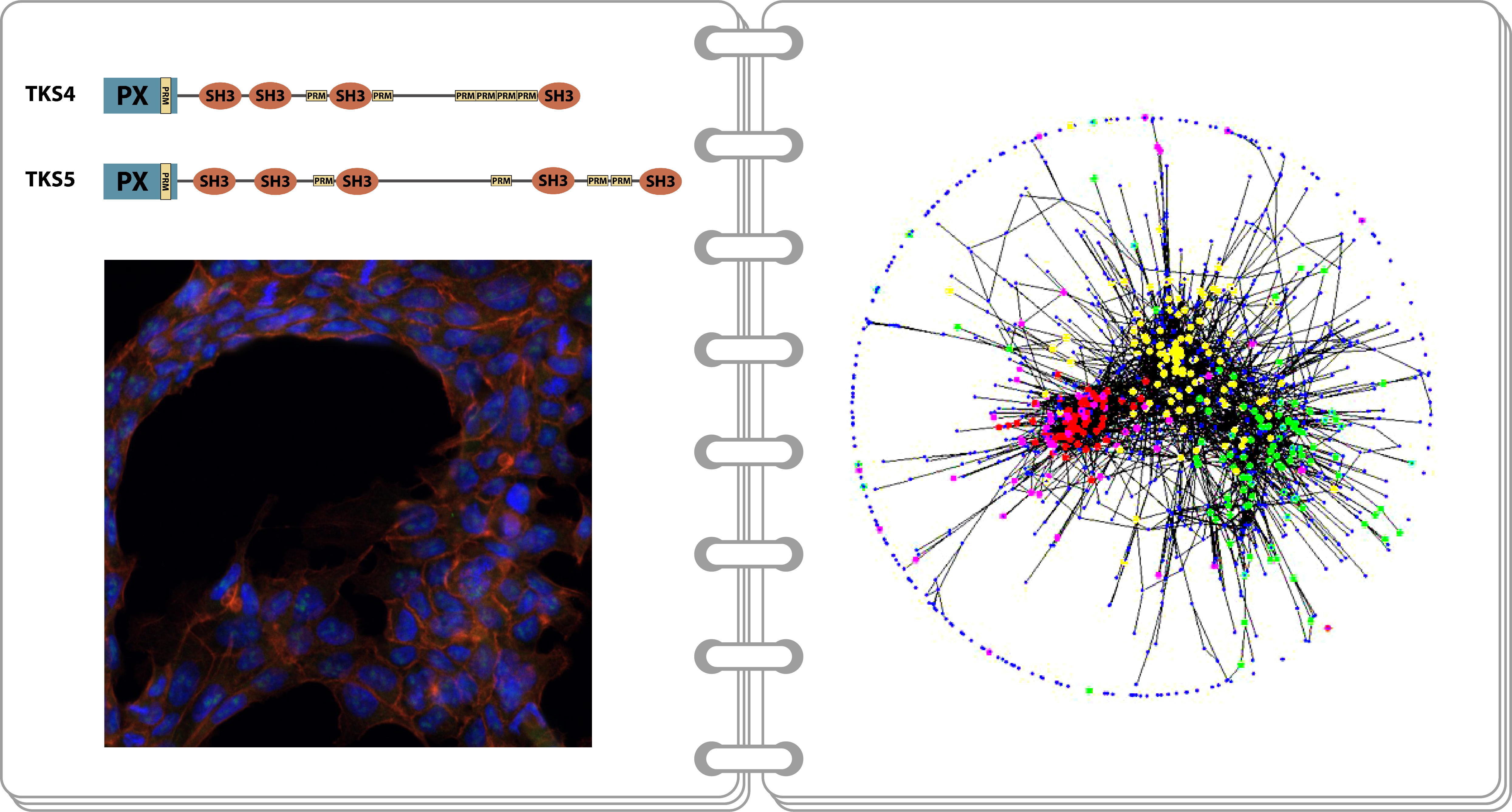
Tks4 and Tks5 are two members of the tyrosine kinase substrate (Tks) family of scaffold proteins. They both possess an N-terminal PX domain and four (Tks4) and five (Tks5) SH3 domains. This domain structure implies that these proteins are able to bind certain lipids and proteins – a characteristic of scaffold proteins. The Tks4 protein and its gene (SH3PXD2B) have important medical importance since mutations in the SH3PXD2B gene cause the hereditary disorder Frank-ter Haar syndrome (FTHS) showing severe developmental abnormalities. The group has demonstrated that both Tks4 and Tks5 take part in EGF signalling [1-3]. They have proven that a point mutation known to affect FTHS patients results in structural damage to the Tks4 protein which then accumulates in aggresomes in the cells [4]. The group has generated a Tks4 knock-out mouse strain and proved that mesenchymal stem cells derived from these mice have a reduced ability to differentiate into adipogenic and osteogenic lineages [5]. This can partially explain the phenotype observed in FTHS patients. On a molecular level, the group has shown that Tks4, which is also a substrate for Src tyrosine kinase, interacts with the SH2 and SH3 domains of Src in EGF-treated cells [6].

[1] Lányi A, Baráth M, Péterfi Z, Bögel G, Orient A, Simon T, Petrovszki E, Kis- Tóth K, Sirokmány G, Rajnavölgyi É, Terhorst C, Buday L and Geiszt M. (2011) The homolog of the Five SH3-Domain Protein (HOFI/SH3PXD2B) Regulates Lamellipodia Formation and Cell Spreading. PLoS ONE 6, e23653
[2] Bögel G, Gujdár A, Geiszt M, Lányi Á, Fekete A, Sipeki S, Downward J and Buday L. (2012) Frank-ter Haar syndrome protein Tks4 regulates epidermal growth factor-dependent cell migration. J Biol Chem 287, 31321-9
[3] Fekete A, Bőgel G, Pesti S, Péterfi Z, Geiszt M, Buday L. (2013) EGF regulates tyrosine phosphorylation and membrane-translocation of the scaffold protein Tks5. J Mol Signal 8, 8
[4] Ádám C, Fekete A, Bőgel G, Németh Z, Tőkési N, Ovádi J, Liliom K, Pesti S, Geiszt M, Buday L. Accumulation of the PX domain mutant Frank-ter Haar syndrome protein Tks4 in aggresomes. (20115) Cell Commun Signal. 2015 13, 33
[5] Dülk M, Kudlik G, Fekete A, Ernszt D, Kvell K, Pongrácz JE, Merő BL, Szeder B, Radnai L, Geiszt M, Csécsy DE, Kovács T, Uher F, Lányi Á, Vas V, Buday L. (2016) The scaffold protein Tks4 is required for the differentiation of mesenchymal stromal cells (MSCs) into adipogenic and osteogenic lineages. Sci Rep. 2016 6, 34280
[6] Dülk M, Szeder B, Glatz G, Merő BL, Koprivanacz K, Kudlik G, Vas V, Sipeki S, Cserkaszky A, Radnai L, Buday L. (2018) EGF Regulates the Interaction of Tks4 with Src through Its SH2 and SH3 Domains. Biochemistry. 2018 57(28), 4186-4196.
- The study of Caskin scaffold proteins
Caskin (CASK-interacting) scaffold proteins are structurally composed of ankyrin-repeats, SAM and SH3 domains, and several proline-rich regions. The structures of Caskin1 and Caskin2 protein differ mostly in the length of their proline-rich regions. Moreover, they have distinct expression patterns: while Caskin1 is solely expressed in the central nervous system, Caskin2 expression can be observed in several tissues and organs. In collaboration with Péter Tompa’s research team, this group has demonstrated that the Caskin1 protein structure is largely disordered. They have also shown that the proline-rich disordered region is functional since it can bind the Abi2 (Abl-interactior 2) scaffold protein through its SH3 domain [1]. The group has established through studying the signalling pathway of the EphB1 receptor tyrosine kinase that the autophosphorylated receptor is able to form a complex with Caskin1 through the Nck linker protein. The receptor-bound Caskin1 is then phosphorylated on the two tyrosine residues found in its SH3 domain [2]. In recent years, the group has created a Caskin1/Caskin2 double knock-out mouse strain and is currently working on its characterization.
[1] Balazs A, Csizmok V, Buday L, Rakacs M, Kiss R, Bokor M, Udupa R, Tompa K, Tompa P. (2009) High levels of structural disorder in scaffold proteins as exemplified by a novel neuronal protein, cask-interactive protein1. FEBS J 276, 4168
[2] Pesti S, Balázs A, Udupa R, Szabó B, Fekete A, Bögel G and Buday L. (2012) Complex formation of EphB1/NCK/Caskin1 leads to tyrosine phosphorylation and structural changes of the Caskin1 SH3 domain. Cell Commun Signal 10, 36
- Tyrosine phosphorylation of SH3 domains: an emerging novel regulatory mechanism of protein-protein interactions
The group, led by László Buday, has been intensively studying the function of SH3 domain in the past 20 years. They investigated several SH3 domain containing proteins, such as Grb2, Crk, Nck, Vav2, cortactin, Lck, SLP-76, Caskin1, Src, and the family of Tks proteins [1-8]. One of their important discoveries is that SH3 domains can be subject of tyrosine phosphorylation in certain situations that may influence protein-protein interactions [9]. Currently, the research group systematically investigates the tyrosine phosphorylation of the SH3 domains and its role in signal transduction.

[1] Buday, L., Khwaja, A., Sipeki, S., Faragó, A., and Downward, J. (1996) Interactions of Cbl with two adaptor proteins, Grb2 and Crk, upon T cell activation. J Biol Chem 271, 6159-6163
[2] Wunderlich, L., Faragó, A., and Buday, L. (1999) Characterization of interactions of Nck with Sos and dynamin. Cellular Signalling 11,25-29
[3] Wunderlich, L., Gohér, A., Faragó, A., Downward, J., and Buday, L. (1999) Requirement of multiple SH3 domains of Nck for ligand binding. Cellular Signalling 11, 253-262
[4] Buday, L. (1999) Membrane-targeting of signalling molecules by SH2/SH3 domain-containinmg adaptor proteins. Biochim Biophys Acta 1422,187-204
[5] Tamas P, Solti Z, Bauer P, Illes A, Sipeki S, Bauer A, Farago A, Downward J, Buday L. (2003) Mechanism of EGF regulation of Vav2, a guanine nucleotide exchange factor for Rac. J Biol Chem 278, 5163-5171
[6] Illes A, Enyedi B, Tamas P, Balazs A, Bogel G, Lukacs M, Buday L. (2006) Cortactin is required for integrin-mediated cell spreading. Immunol Letters 104, 124-130
[7] Buday, L., Downwrad, J. (2007) Roles of cortactin in tumor pathogenesis. BBA – Reviews on cancer 1775, 263-273
[8] Hem CD, Sundvold-Gjerstad V, Granum S, Koll L, Abrahamsen G, Buday L, Spurkland A. (2015) T cell specific adaptor protein (TSAd) promotes interaction of Nck with Lck and SLP-76 in T cells. Cell Commun Signal 13, 31
[9] Pesti S, Balázs A, Udupa R, Szabó B, Fekete A, Bögel G and Buday L. (2012) Complex formation of EphB1/NCK/Caskin1 leads to tyrosine phosphorylation and structural changes of the Caskin1 SH3 domain. Cell Commun Signal 10, 36
- Genome Annotation
In the 21st century medical sciences, drug development, agriculture, biotechnology as well as evolutionary biology rely increasingly on information originating from genome projects.
A crucial step in the interpretation of genome sequences is the identification of protein-coding genes and prediction of their structure with bioinformatic tools. The success of most subsequent steps of biological research exploiting genomic sequences depends on the quality of these data. The difficulties of gene identification may be illustrated by the fact that – despite significant improvements in gene-prediction technologies – prediction of the structure of protein-coding genes is still unreliable: according to current estimates, the structure of less than 50% of predicted genes is correct [1].
To solve this problem we have developed a method that helps to decide whether an experimentally determined or in silico predicted protein-coding sequence is erroneous (abnormal, incomplete, mispredicted). The MisPred approach is based on the principle that a protein-coding gene is likely to be mispredicted if some of its features (or features of the protein it encodes) conflict with our current knowledge about protein-coding genes and proteins [2-4].
[3] Nagy A, Hegyi H, Farkas K, Tordai H, Kozma E, Bányai L and Patthy L (2008) Quality control of gene predictions. In: Frishman D., Valencia A. (eds) Modern Genome Annotation. Springer, Vienna.
Identification of erroneous genes/proteins is of crucial importance since it may protect users from drawing erroneous conclusions based on erroneous data [5-7].
Nevertheless, it is also important to correct the errors of sequences. Given the heavy contamination of sequence databases with incorrect sequences, identification of erroneous entries and their correction may be performed only by automated quality control and correction protocols. In our FixPred project we developed tools for the automatic correction of erroneous sequences [8-9].
The next step of genome annotation is the definition/prediction of all aspects of the molecular functions and biological roles of the various genes/proteins in health and disease. This is essential if we wish to assess the relevance of each gene for medicine, agriculture or biotechnology. We have performed complex bioinformatic analyses to predict the structure and function of novel proteins, primarily from Metazoa. These analyses include assignment of the novel proteins/domains to known domain-families, prediction of their most probable function, predictions of the subcellular localization and structure-function aspects of the proteins.
In the course of such analyses we have characterized some novel multidomain proteins (e.g. WFIKKN1, WFIKKN2) [10-11] and have defined several novel protein domain-types (e.g. the NTR-domains [12], PAN-domains [13], WIF-domains [14], LCCL-domains [15]).
[14] Patthy L (2000) The WIF module. Trends Biochem Sci 25(1):12-13.
[15] Trexler M, Bányai L and Patthy L (2000) The LCCL module. Eur J Biochem 267(18):5751-5757.
In collaboration with the NMR group of Gottfried Otting (Research School of Chemistry, The Australian National University, Canberra, Australia) we were the first to solve the structure of an LCCL-domain [16], a NTR-domain [17] and an WIF-domain [18], confirming our structure predictions.
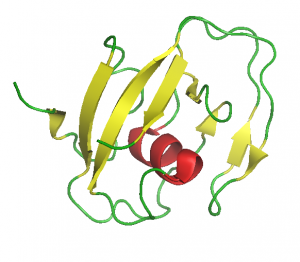
LCCL-domain (1JBI.pdb)
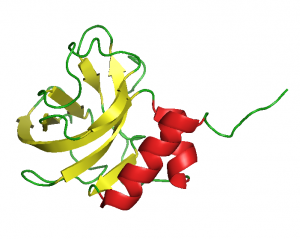
NTR-domain (1UAP.pdb)
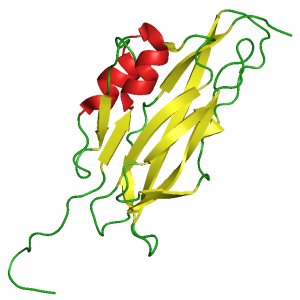
WIF-domain (2D3J.pdb)
Based on the analysis of genomes, transcriptomes and proteomes of various organisms, we have provided evidence that exon-shuffling played a key role in the evolution of the proteins that are indispensable for the multicellular body plan of Metazoa [19-21].
Based on comparison of human genomes from normal and tumor tissues we have developed a procedure for the identification of ‘cancer genes’ that play important roles in tumor evolution [22].
- Studies on some medically important multidomain proteins involved in regulating signal transduction
In our experimental work we have been focusing on the function of the multidomain proteins WFIKKN1, WFIKKN2 and Wnt Inhibitory Factor 1.
We have shown that both WFIKKN proteins are potent inhibitors of GDF8 (myostatin) and the closely related GDF11, but do not block the signaling activity of other members of the TGFβ family [1-5].
The medical importance of WFIKKN1 and WFIKKN2 (and/or their derivatives) is that they have the potential to be used as antimyostatic agents in the therapy of muscle wasting diseases. In the framework of the National Program for Protein Science, Hungary we are developing molecules that can serve as more potent and more specific myostatin antagonists.
Wnt Inhibitory Factor 1 protein is also of major medical interest since epigenetic silencing of the WIF1 gene (and the resulting abnormal activation of the Wnt- β catenin signaling pathway) is associated with many types of cancer. Recombinant WIF1 protein blocks tumor progression, however, the potency of WIF1 protein as a Wnt antagonist differs markedly for various types of Wnts [8-9]. Another problem with the use of WIF1 as a Wnt-antagonist is that the protein may also be involved in the regulation of the hedgehog pathway [10].
In the framework of the National Program for Protein Science, Hungary we are producing WIF1 mutants that bind to the various Wnts with higher specificity, making them more promising antitumor agents.
Since the WIF1 gene is silenced in a variety of tumors, expression of the WIF1 protein is a valuable biomarker in tumor diagnosis and precision tumor therapy. In the framework of the ELTE FIEK project for Biotechnology, Hungary we are developing a sensitive and reliable QPCR-based protocol to quantitate expression of the WIF1 gene in various tumor tissues.
Lab equipments, services
Tools
Cell culture laboratory
Excella ECO-170 CO2 incubator
ESCO CLASS II BSC incubator
NÜVE ES110 incubator
Zeiss AXIO phase contrast microscope
Protein purification and characterization
CARY 300 UV-VIS spectrophotometer
CARY Eclipse spectrofluotimeter
UNICORN FPLC equipment
ChemiImager 5500 gel-documentation systyem
LSM710 Confocal Microscope
Leader
László Buday
Members
| Name | Status | Phone | Group | Room | |
| László Bányai | tudományos főmunkatárs | +36 1 3826 749, 751 | Signal Transduction and Functional Genomics Research Group | D3.13A | |
| Ildikó Bódi Pálné | laboratóriumi asszisztens | +36 1 3826 749 | Signal Transduction and Functional Genomics Research Group | D3.12A | |
| László Buday | professzor | +36 1 3826 701 | Signal Transduction and Functional Genomics Research Group | D3.01A | |
| Metta Dülk | tudományos segédmunkatárs | +36 1 3826 706 | Signal Transduction and Functional Genomics Research Group | D3.03A | |
| Dóra Kállai | szakdolgozó | +36 1 3826 706 | Signal Transduction and Functional Genomics Research Group | D3.03A | |
| Krisztina Kerekes | tudományos segédmunkatárs | +36 1 3826 752 | Signal Transduction and Functional Genomics Research Group | D3.13B | |
| Kitti Koprivanacz | tudományos segédmunkatárs | +36 1 3826 726 | Signal Transduction and Functional Genomics Research Group | D3.12 | |
| Gyöngyi Kudlik | tudományos munkatárs | +36 1 3826 706 | Signal Transduction and Functional Genomics Research Group | D3.03A | |
| Balázs Merő | tudományos segédmunkatárs | +36 1 3826 726 | Signal Transduction and Functional Genomics Research Group | D3.12 | |
| László Patthy | professzor emeritus | +36 1 3826 751 | Signal Transduction and Functional Genomics Research Group | D3.13A | |
| Zita Solti | laboratóriumi asszisztens | +36 1 3826 781 | Signal Transduction and Functional Genomics Research Group | Ny3.06-07 | |
| Bálint Szeder | tudományos segédmunkatárs | +36 1 3826 726 | Signal Transduction and Functional Genomics Research Group | D3.12 | |
| Mária Trexler | tudományos főmunkatárs | +36 1 3826 751 | Signal Transduction and Functional Genomics Research Group | D3.13A | |
| Virág Vas | tudományos munkatárs | +36 1 3829 706 | Signal Transduction and Functional Genomics Research Group | D3.03A |